|
Native proteins must be in an unfolded
conformation during degradation and stabilizing a substrate
protein against unfolding can prevent its proteolysis. ATP-dependent
proteases can accelerate the denaturation of their substrates.
It has been shown, for example, ClpAP can unfold green fluorescent
protein (GFP) at a rate that is six-orders of magnitude
faster than the rate at which the protein would unfold spontaneously.
Subsequent studies have found that the proteasome,
ClpXP, Lon and the archebacterial proteasome can also actively
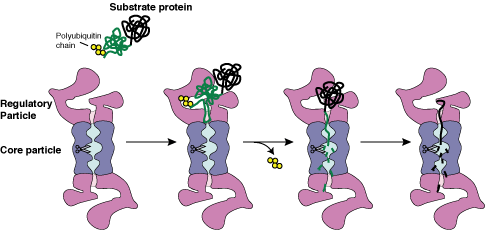
unfold their substrates. We have found that the proteasome
catalyzes unfolding by engaging the substrates through their
unstructured initiation sites and sequentially unraveling
them from their degradation initiation sites.
In a multidomain protein, the domain closest
to the initiation site is degraded first, whereas domains
located further away are degraded subsequently. Stabilizing
a domain close to the initiation site protects downstream
domains from degradation but not vice versa. Experiments using
fluorescence resonance energy transfer (FRET) confirms that
the bacterial protease ClpAP translocates its substrate sequentially
to its proteolytic sites. ClpXP and FtsH have also been shown
to degrade their substrates sequentially along the polypeptide
chain.
|
|
Sequential
unfolding and degradation by the proteasome
|
|

courtesy Neil Jaffe
|
|


