Research Areas
Biological Methane Oxidation
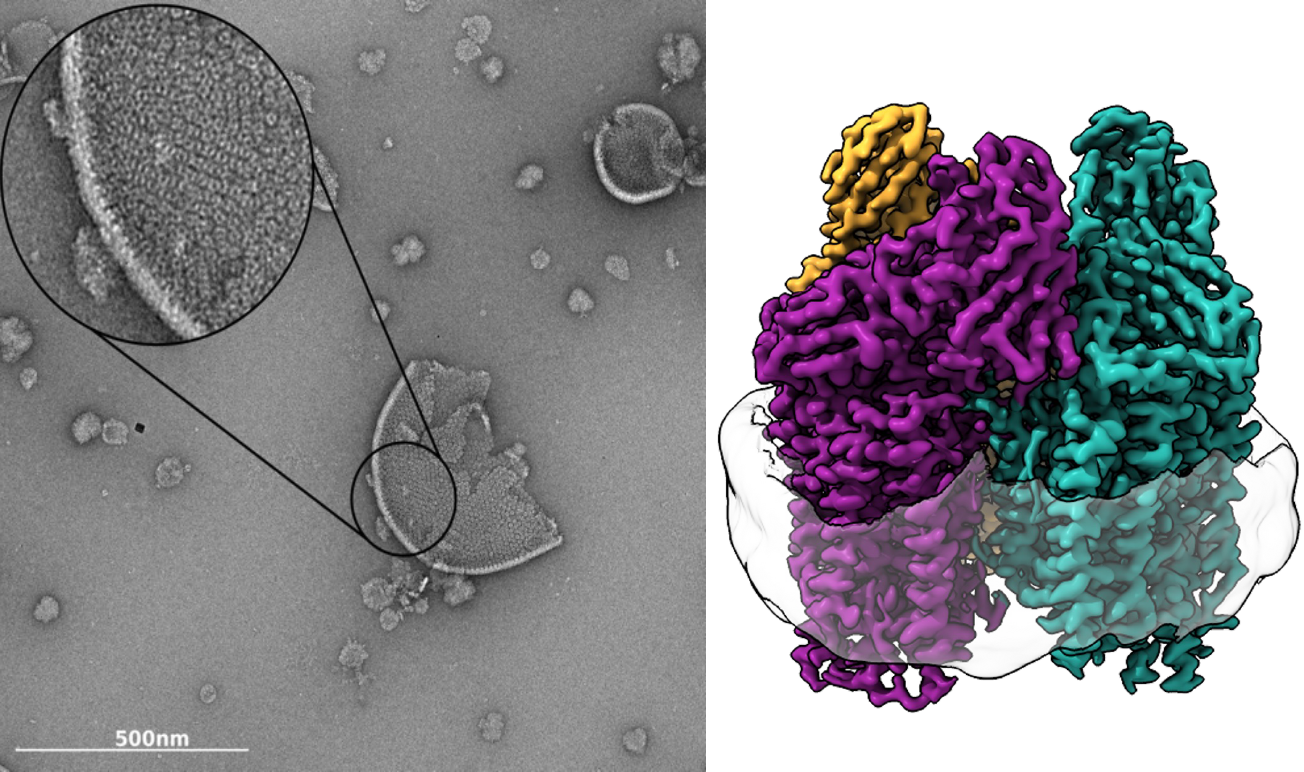
Left, Negative stain micrograph of Methylococcus capsulatus (Bath) isolated membranes. Tightly packed pMMO trimers are clearly visible in the membrane. Right, 2.14 Å resolution cryoEM map of M. capsulatus (Bath) pMMO showing the trimer and the encircling nanodisc. Symmetrical αβγ protomers are colored in purple, teal, and gold.
Methanotrophic bacteria oxidize methane to methanol in the first step of their metabolic pathway. Whereas current catalysts that can selectively activate the 105 kcal mol-1 C-H bond in methane require high temperatures and pressures, methanotrophs perform this chemistry under ambient conditions using methane monooxygenase (MMO) enzymes. The primary MMO in nature, particulate MMO (pMMO), is a th ree-subunit, integral membrane protein. Despite extensive research and the availability of multiple crystal structures, the active site structure and chemical mechanism of pMMO remain one of the major unsolved problems in bioinorganic chemistry. Current efforts in the laboratory are directed at elucidating the atomic details of the copper active site, understanding the mechanisms of dioxygen activation and methane oxidation, including how substrates, products, electrons, and protons access the active site, and probing the function of pMMO within the larger context of methanotroph physiology.
To learn more about this project:
Biosynthesis and Transport of Methanobactin
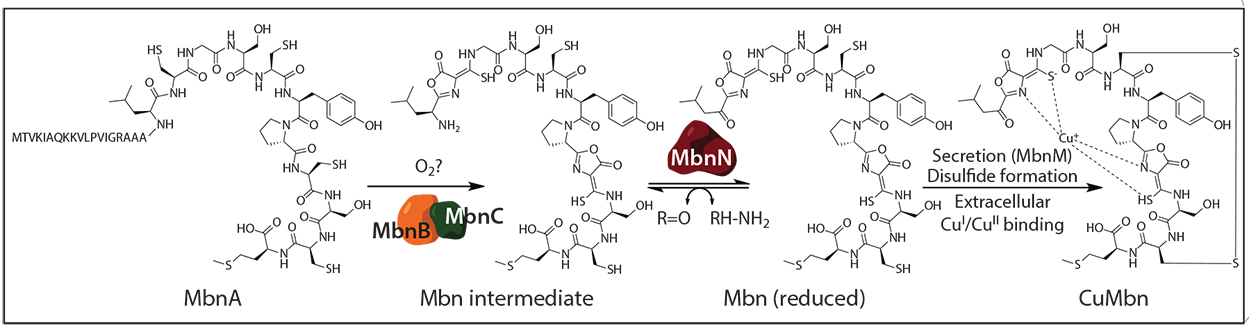
Biosynthesis of Methylosinus trichosporium OB3b Mbn from its precursor peptide MbnA.
Copper acquisition is particularly important for methanotrophs because their primary metabolic enzyme, particulate methane monooxygenase (pMMO), requires copper for activity. Some methanotrophs meet their high requirement for copper by secreting and reinternalizing methanobactin (Mbn), a peptide-derived, copper-chelating natural product. Beyond its role in methanotrophy, Mbn is a potential therapeutic for Wilson disease, a genetic disorder of toxic copper overload.
All Mbns characterized thus far bind Cu(I) with two nitrogen-containing heterocycles and two adjacent thioamide groups. Mbn is biosynthesized by post-translational modification of a precursor peptide called MbnA, and bioinformatics analyses revealed that the mbnA gene is located within an operon that also includes genes encoding machinery for Mbn transport and biosynthesis. Importantly, these operons are found in a range of non-methanotrophic bacteria, including gram-positive pathogens, suggesting the Mbn-like natural products may be widespread in nature. Current efforts in the laboratory are focused on discovering new Mbns and related natural products, characterizing the proteins involved in Mbn export and import, and unraveling the mechanisms of its biosynthesis.
To learn more about this project:
Metal Transport
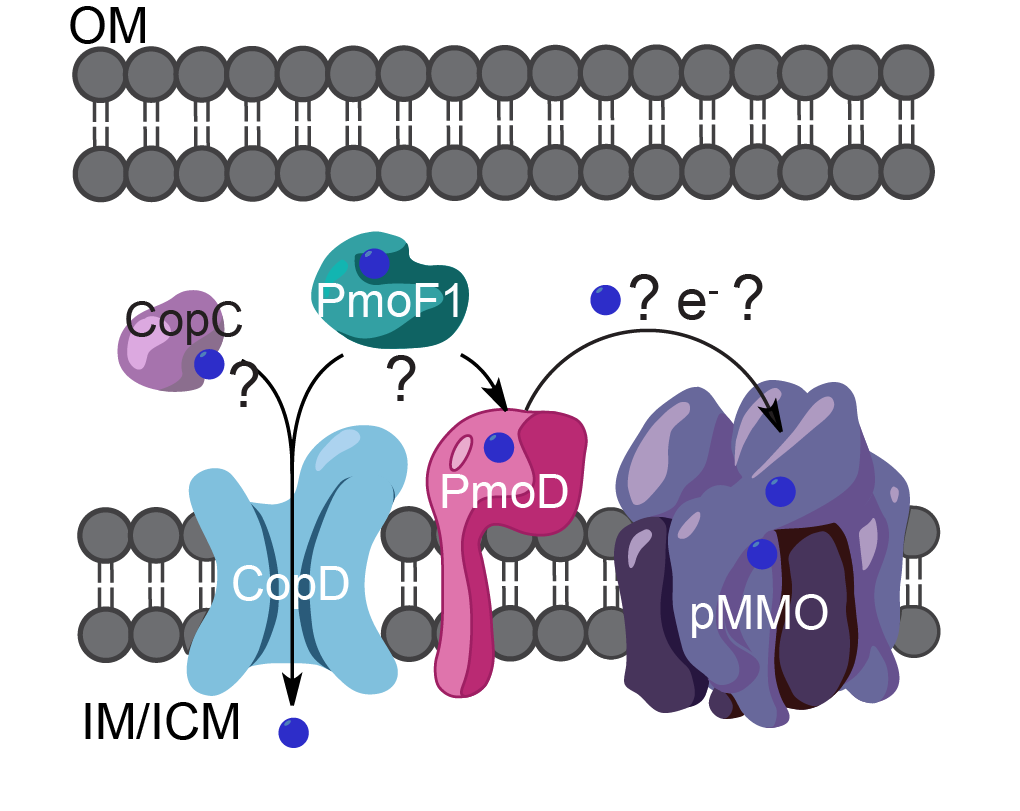
Predicted cellular locations of copper binding proteins PmoD, CopC, CopD, and PmoF1 (OM, outer membrane; IM, inner membrane; ICM, intracytoplasmic membrane). Copper ions are shown as blue spheres. The arrows and question marks indicate possible functional relationships between the proteins.
Metalloproteins comprise close to one third of all proteins and almost half of all enzymes. Acquisition and management of metal ions is therefore a critical part of metabolism for all forms of life. Metals must be handled such that the correct ions are provided to essential enzymes and proteins, but do not accumulate to deleterious levels. Membrane transporters, metallochaperones, and metal sensors maintain metal ion concentrations in cells and cellular compartments, and aberrant handling of metal ions is linked to numerous human diseases. Understanding metal homeostasis on the molecular level is thus a central problem in cell biology and bioinorganic chemistry.
Current efforts in the laboratory are focused on bacterial copper binding proteins, including members of the CopC, CopD, PCuAC, and PmoD families, of which the latter family is found only in methane and ammonia oxidizing bacteria. Current efforts in the laboratory are directed at determining the functions and copper-binding properties of these proteins.
To learn more about this project:
