| Symbol: Symbol: Su(fu) {Links}
|
Flybase ID: {Flybase_ID} |
| Synonyms: {Name}
|
{GadFly} |
| Function: {Short_Function} |
{LocusLink} |
| Keywords: {Keywords} |
{Interactive_Fly} |
{Summary}
|
| {Function/Pathway}{Function}
|
- Class 0, I, and II fused phenotype is suppressed by Su(fu)LP (class
II has a cos2 phenotype in addition) (Preat,
1993)
- mutations that eliminate Su(fu) did not increase the minimal induction
of ptc observed in anterior sgg mutant clones (Price,
2002), however others show that ptc expression and wing duplications
increase in a Su(fu) mutant background (Jia,
2002).
- In the absence of Su(fu) protein, inhibiting
the PKA-dependent phosphorylation of Ci now results in the nuclear import
of full-length Ci (Methot,
2000), implying that Su(fu) also acts to retain Ci in the cytoplasm.
Su(fu) has been shown to bind directly to the N-terminal region of Ci
in yeast two-hybrid assays ([Monnier,
1998), but biochemical characterization of the proteins in vivo
suggest that this interaction results in the formation of a complex
distinct from that associated with microtubules (Stegman et al. 2000).
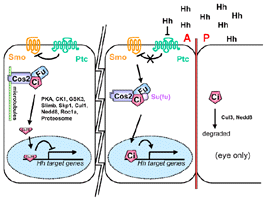
This implies that Su(fu) may act in concert with Cos-2 to sequester
Ci in the cytoplasm after it has been released from microtubules [Ingham,
2001]
|
- Su(fu) blocks Ci nuclear import through the N-term region of Ci (Wang,
2000)
- Fu’s aa 306-436 was sufficient for binding to Su(fu) (Y2hybrid
and GST) (Monnier,
1998)
- Ci’s aa 1-346 was sufficient for binding to Su(fu) (Y2hybrid
and GST) (Monnier,
1998)
- binds to Fu and Ci (Y2hybrid, GST-pull down, co-IP) (Monnier,
1998)
- S: may form a stabilizing bridge
- by genetics has been shown to antagonize fu in a dose-dependent manner
- is found in a complex with b-catenin and functions as a negative regulator
of Tcf dependent transcription (Meng.
2001)
- This interaction is most likely in the C-term of Su(fu)(Meng.
2001)
- overexpression of Su(fu) in cell culture leads to a reduction in nuclear
b-cateinin and Tcf-dependent transcription (Meng.
2001)
- This is dependent on LMB, thus implicating a CRM1 dependent mechanism
(Meng.
2001)
- hSu(fu) increases Gli1 and Gli3 DNA binding in vitro (Pearse,
1999)
- mSu(fu) interacts with SAP18, a component of the mSin3 and histone
deacetylase complex. (Cheng,
2002)
- mSu(fu) functionally cooperates with SAP18 to repress transcription
by recruiting the SAP18-mSin3 complex to promoters containing the Gli-binding
element. (Cheng,
2002)
- maximal response of cells to Hh signaling requires the dissociation
of Ci from Su(fu)
- Su(fu) may act in concert with Cos-2 to sequester Ci in the cytoplasm
after it has been released from microtubules
- Su(fu) can enter the nucleus bound to Ci and attenuate its transcription-activating
activity, a view supported by the properties of the vertebrate Su(fu)
ortholog (Kogerman,
1999).
|
- Su(fu) mRNA is ubiquitous throughout embryonic development (Pham,
1995)
- Drosophila Su(fu) is in cytoplasm and Chick Su(fu) is found in the
nucleus (Pearse,
1999)
|
- animals homozygous for a deletion of the gene are fully viable and
fertile (Préat,
1992)
- In a cos2 Su(fu) double mutant en is ectopically expressed (Wang,
2000)
- Mutations cause an inc. in the expression of Hh-target genes in a
dose-dependent manner while simultaneously reducing [Ci-155] by some
mechanism other than proteolysis to Ci-75 (Ohlmeyer,
1998 )
- Mutations inc. the effects of ptc and PKA mutant clones (Ohlmeyer,
1998 )
|