      |
   |
Detection of foreign RNA in the cytoplasm-
One group of intracellular responders, the RIG-I like receptors (RLRs), includes proteins encoded by the retinoic acid-inducible gene I (RIG-I), the melanoma differentiation-associated gene 5 (MDA5), and the laboratory of genetics and physiology gene 2 (LGP2). Similar in structure and function, these proteins share significant sequence homologies that define them as an evolutionarily conserved gene family (Figure 1A).
The RLR proteins are thought to share the ability to detect molecular signatures of virus infection and activate antiviral signaling cascades, but differ in both their RNA recognition capacities and signaling properties. RIG-I, MDA5, and LGP2, share homologous DECH-box helicase domain regions that have intrinsic dsRNA binding and ATP hydrolysis functions, and a C-terminal domain that has been implicated in binding to RNA termini and auto-regulation. RIG-I and MDA5 both have tandem N-terminal caspase activation and recruitment domain (CARD) motifs, protein interaction domains that mediate associations with upstream and downstream regulatory machinery. The CARDs are regulated by post-translational modifications, including ubiquitination and phosphorylation. Current evidence indicates that RNA recognition by RIG-I and MDA5 is accompanied by CARD dephosphorylation, enabling their productive interaction with signaling machinery. Much of the RLR signaling pathway was developed by detailed study of RIG-I, the prototype RLR, and investigations of MDA5 have contributed greatly to a general paradigm for RLR signaling (Figure 1B).
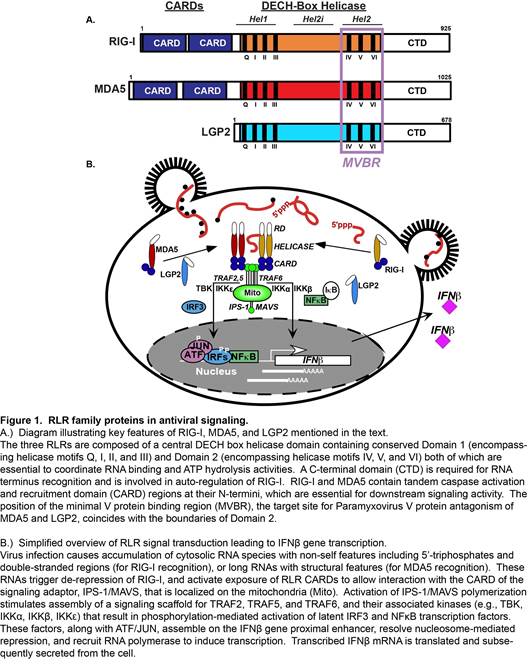
Figure 1
Interaction with non-self ligand RNAs leads to exposure of active RLR CARDs, enabling association with the CARD of an essential mitochondrial antiviral signaling protein, MAVS. MAVS is thought to act as a polymeric signaling scaffold that facilitates assembly and activation of signaling proteins including TRAF2, TRAF5, and TRAF6, and associated serine kinases, including IKK a , IKK b , IKK g , IKK e , and TBK1. These kinases activate the master transcription regulators, IRF3 and NF k B that induce production of IFN b and diverse antiviral target genes. Secreted IFN b amplifies the antiviral response via IFNAR-JAK-STAT signaling, inducing the expression of many more effectors that together produce a cellular antiviral state that provides a broadly effective barrier against virus replication.
While MDA5 has been the subject of many investigations, its redundant and non-redundant functions with respect to RIG-I have only recently been unraveled. The precise features of an MDA5-specific non-self RNA ligand have not been revealed, contributing to a relative deficiency in mechanistic details regarding the basis for MDA5 non-self RNA recognition leading to signal transduction. For LGP2, the absence of CARDs or other recognizable signaling domains has impeded drawing clear comparisons with the functions of the other RLR proteins. Furthermore, the apparently antithetical reported functions of LGP2 as both a positive and negative regulator of antiviral responses have been difficult to reconcile. These mechanistic details leave a large gap in our understanding of RLR signaling, and contribute to contradictory reports that implicate MDA5 and LGP2 as either collaborators or competitors in the antiviral system. The complex relationship of MDA5 and LGP2 is a recent focus of our research, and a unifying model has been developed to clarify their roles in the regulation of antiviral signaling (Figure 2).
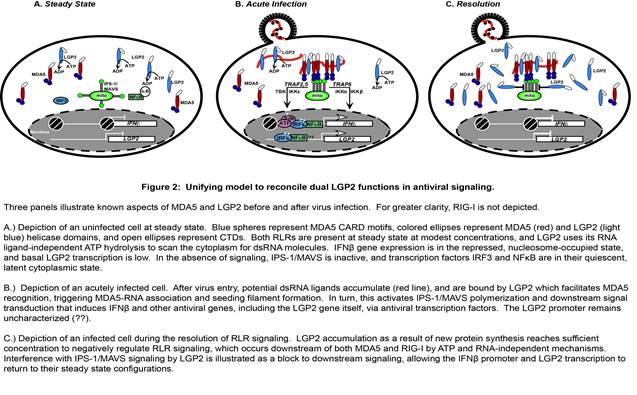
Figure 2
Titrating LGP2 expression into MDA5-dependent signaling demonstrates that low levels of LGP2 are synergistic with MDA5, a process that requires LGP2 ATP hydrolysis and RNA binding activities. The MDA5-stimulating activity of LGP2 is revealed in a narrow stoichiometric range, and further titration of LGP2 expression ultimately achieves an inhibitory concentration that disrupts RLR signaling activity.
Although many aspects of MDA5-LGP2 synergy remain to be addressed experimentally, our recent work has unraveled one element of this co-regulation (Figure 3). It was observed that LGP2 catalytically increases the initial rate and stability of MDA5-dsRNA interactions. In addition, although LGP2 did not form dsRNA filaments by itself, the presence of LGP2 dramatically altered the quantity and quality of MDA5 filaments. LGP2 enabled MDA5 to generate a greater number of filaments, although the filaments were shorter in length. LGP2 enhances MDA5 activity by facilitating its initial RNA interaction, and has the effect of increasing the number and regulating the length of MDA5-RNA filaments. This MDA5 co-activation requires LGP2 ATP hydrolysis and RNA-binding activities, as well as an intact MDA5 oligomeric interface. This phenomenon apparently ensures MDA5 RNA recognition and optimizes signaling output, providing a plausible mechanistic basis for LGP2 co-activation.
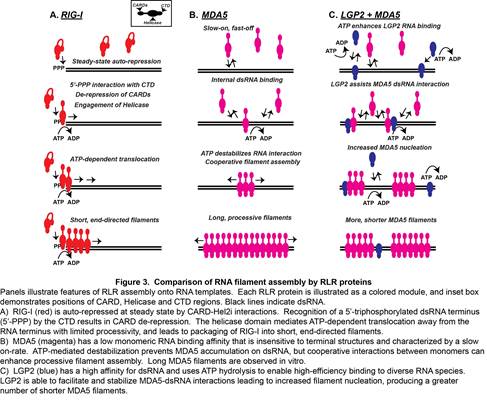
Figure 3
|
 |

