|
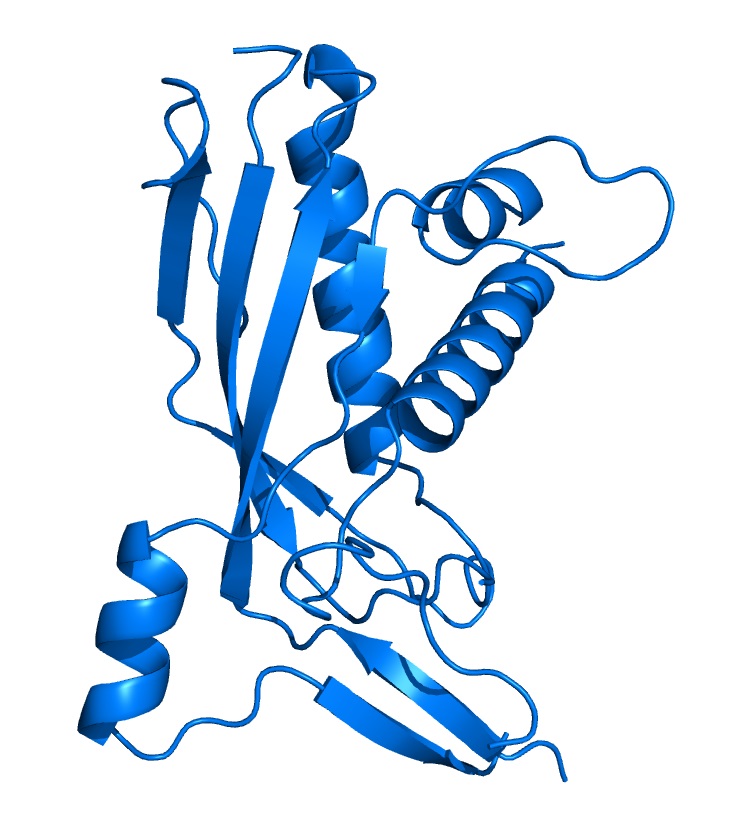
1) They are responsible for maintaining the topological state of DNA and are involved in a variety of crucial cellular processes. 2) Their involvement in key processes has lead to the development of drugs whose targets are topoisomerases. 3) Topoisomerases catalyze a complex reaction that involves cutting and resealing the DNA and passing DNA strands through this break. These reactions are not easy to visualize or understand. The structures of several topoisomerases and fragments of them have truly led to a near-atomic picture of the way a very complex reaction is catalyzed. 4) Topoisomerases are excellent examples of complex molecular machines that perform a complicated reaction in the cell. Type I enzymes work in the absence of an external energy source, such as ATP, and for this reason present an opportunity to understand a process where the energy to drive large domain movements is harnessed from the energy stored in the DNA. 5) The structural studies may provide the information to develop new chemotherapeutic agents. In the last few years our laboratory has worked on the structure of
several different type I topoisomerases, including E. coli DNA
topoisomerases I and III (type IA), and vaccinia virus and Deinococcus
radiodurans topoisomerase I (type IB), and more recently on Methanopyrus
kandleri topoisomerase V as well as Streptococcus pneumoniae. In all
cases, a combination of structural and biochemical work has helped elucidate
the atomic basis of the catalytic mechanism of these enzymes. |
|
|
|
|
|
|
|
|
|
|
|
RNase P is one of only two ribozymes conserved in all three kingdoms of life and is required in the 5’ maturation of all tRNAs. In the last years, we solved the crystal structures of the specificity domain of Bacillus subtilis and Thermus thermophilus RNase P and also of the intact RNA component of T. maritima RNase P. In the structure of the intact molecule, the entire RNA catalytic component is revealed, as well as the arrangement of the two structural domains. The structure shows the general architecture of the RNA molecule, the inter- and intra-domain interactions, the location of the universally conserved regions, the regions involved in pre-tRNA recognition, and the location of the active site. A model with bound tRNA is in excellent agreement with all existing data and suggests the general basis for RNA-RNA recognition by this ribozyme. This is the first structure of an A-type bacterial RNase P solved and represents one of the largest RNA molecules whose structure is known. We have also obtained structures of the holoenzyme and a tertiary complex involving the RNA component, the protein component, and pre-tRNA. In the future we plan to continue our work on RNase P by investigating the specificity of the active site on a structural basis |
|
|
|
|
|
|
|
|
|
|
|
Other Proteins and Protein-Nucleic Acid Complexes
Additional projects in the lab concern individual proteins and protein-nucleic acid complexes found in various bacteria and archaea. One of these projects is the elucidation of a Type III-A CRISPR effector complex in Staphylococcus epidermidis. This complex is composed of multiple protein subunits in different stoichiometries and a guide RNA known as a CRISPR RNA (crRNA). Type III CRISPR effector complexes are unique in the CRISPR world for their ability to target and cleave both RNA and DNA through multiple mechanisms. Our lab is focused on determining the structure of the effector complex to better understand the quaternary organization of the complex as well as the structural dynamics that occur during its interference function.
|
|
|
|
|
|
|
|
|
|
|
|
Last updated: 6/17/2019 |
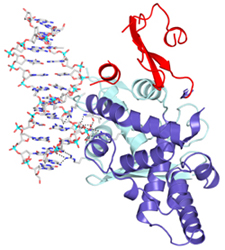 The long term goal of our
work is to understand the catalytic mechanism of these molecules in atomic
detail. In particular, we are interested in understanding how these enzymes
perform complex topological rearrangements of DNA molecules. DNA
topoisomerases are of interest for several reasons:
The long term goal of our
work is to understand the catalytic mechanism of these molecules in atomic
detail. In particular, we are interested in understanding how these enzymes
perform complex topological rearrangements of DNA molecules. DNA
topoisomerases are of interest for several reasons: 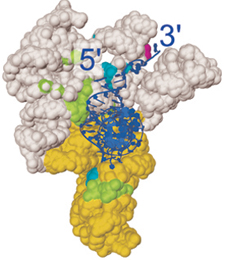 A second large research
area is the structure of large RNA molecules and in particular RNase P RNA
plays a pivotal role in biology as it is involved in many cellular processes.
It is also unique amongst nucleic acids in being able to perform chemical
catalysis. In the cell, RNA molecules can self-cleave or process other RNA
molecules in a manner that up to a few years ago was completely unknown and
unexpected. These findings have led to the idea of a pre-biotic RNA-world,
where RNA molecules were the first molecules to appear. The discovery of the
catalytic properties of RNA molecules has also rekindled the interest on
these molecules both from a basic and from an applied point of view.
A second large research
area is the structure of large RNA molecules and in particular RNase P RNA
plays a pivotal role in biology as it is involved in many cellular processes.
It is also unique amongst nucleic acids in being able to perform chemical
catalysis. In the cell, RNA molecules can self-cleave or process other RNA
molecules in a manner that up to a few years ago was completely unknown and
unexpected. These findings have led to the idea of a pre-biotic RNA-world,
where RNA molecules were the first molecules to appear. The discovery of the
catalytic properties of RNA molecules has also rekindled the interest on
these molecules both from a basic and from an applied point of view.