Biographical
Sketch
Robert A. Lamb, Ph.D., Sc.D.--
John Evans Professor of Molecular and Cellular Biology in the Department of Molecular
Biosciences at Northwestern University, Professor of Microbiology-Immunology
at Northwestern University Medical School and an Investigator of the Howard Hughes
Medical Institute.
Dr. Lamb received his undergraduate degree reading biochemistry at the University of Birmingham, England, and he received his Ph.D. and Sc.D. degrees from the University of Cambridge. He came to the United States to do postdoctoral work with Purnell Choppin at the Rockefeller University, where he later became a faculty member before joining the faculty of Northwestern University. His honors include consecutive NIH MERIT awards. He is past president of the American Society for Virology. Dr. Lamb is a member of the National Academy of Sciences and a Fellow of the American Academy of Arts and Sciences.
Contact Information
Tel: (847)-491-5433
Fax: (847-491-2467
e-mail: ralamb@northwestern.edu
Dr. Lamb's CV
Links (Research-Related,
Journals, Funding, Grants)
Short Summary of Research
Robert Lamb's laboratory studies the replication of influenza virus and
paramyxoviruses, particularly virus-mediated membrane fusion, the action
of the M2 and BM2 protein-selective ion channels, enveloped virus assembly
and how these viruses defeat innate immunity.
Structure and Replication of Influenza Virus
and Paramyxoviruses
Our laboratory is investigating the molecular structure and the mechanism
of replication of influenza A, B and C viruses and the paramyxovirus SV5
(simian virus 5). Influenza viruses cause important diseases in humans
and animals. Influenza A and B viruses have tremendous socioeconomic consequences,
for influenza continues to occur in regular epidemics and occasional pandemics
and is a leading cause of morbidity and mortality. Paramyxoviruses cause
many biologically and economically important diseases of humans and lower
animals. Besides SV5, these viruses include human parainfluenza virus
types 1-4, mumps virus, measles virus, canine distemper virus, Newcastle
disease virus of chickens, and rinderpest of cattle.
Influenza virus and SV5 are being studied not only because of their importance as the causative agents of major diseases but also because they provide excellent models for a variety of integral membrane proteins, particularly the processes by which these viruses enter cells and assemble at the plasma membrane. Since the major glycoproteins are also the predominant antigenic determinants of the viruses, knowledge about their structure should enhance our understanding of how they act as immunological targets, thus aiding in developing new vaccines.
|
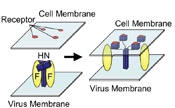
Click Image for Figure and Legend |
Virus Mediated Cell-to-Cell
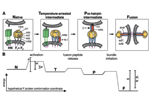
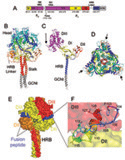
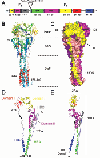
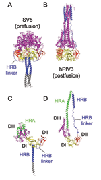
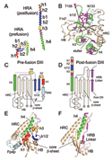
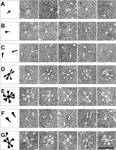
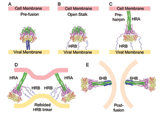
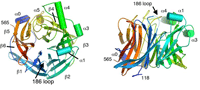
Fusion We have hypothesized that the F protein is a biological nanomachine that undergoes a series of conformational changes and it is controlled such that fusion only occurs at the right time and in the right place. Recently we determined the crystal structure of both the pre-fusion and post-fusion forms of the paramyxovirus F protein in collaboration with Theodore Jardetzky (Northwestern University). The pre-fusion F structure was solved for PIV5 and the post-fusion F structure was solved for hPIV3. In the large part the structures are thought to be representative of the F structures of all paramyxoviruses. To produce a soluble form of the pre-fusion F protein we appended an engineered, trimeric coiled coil domain (GCNt) to heptad repeat B (HRB) to mimic the transmembrane domains. The resulting F protein (F-GCNt) assembled into well-defined trimers. The pre-fusion F stucture contains a globular head attached to a trimeric coiled-coil stalk formed by the C-terminal heptad repeat B region (HRB). The globular head contains three domains (DI-DIII). The fusion peptides at the N-terminus of heptad repeat A (HRA) region are sequestered between adjacent subunits, with cleavage/activation sites exposed at the protein surface. The post-fusion F forms a trimer, which reveals a globular, predominantly beta-sheet containing head domain, a neck region formed by both b-sheet and a-helices and a stalk region that is predominantly a-helical. The structure contained the 6HB expected of the post-fusion conformation of the protein. The only part of the structure lacking electron density are amino acids (95-135) begin 5 residues N-terminal to the internal cleavage site, extending through the fusion peptide (starting at residue 110) and the N-terminal part of HRA , but these residues would be draped flexibly on the exterior of the stalk region. It had been widely anticipated that cleavage of F at the cleavage site was a requirement for conversion to the post-fusion form. Nonetheless, many lines of evidence suggested that the observed F conformation represented the post-fusion form, although the polypeptide chains were intact in the crystal and the fusion peptide was not located at the appropriate end of the 6HB. The PIV5 pre-fusion F and hPIV3 post-fusion F structures are in strikingly different conformations, consistent with a transition from pre- to post-fusion forms. We have observed related forms of the F protein in electron micrographs of F. None of the intersubunit contacts are conserved in the pre- and post-fusion forms. The two F structures are related by flipping the stalk and TM domains relative to the F head. Substantial compacting of the head is observed in hPIV3 post-fusion F compared to PIV5 pre-fusion F. DI domains pivot slightly inwards, shearing intersubunit contacts, and DII domains swing across, contacting neighboring subunits. Individual DI and DII domains in the two structures remain similar. DIII undergoes major refolding between the two structures, projecting a new coiled coil (HRA) upwards and away from DI, the pre-fusion stalk and the viral membrane. The fusion peptide, located at the top of the HRA coiled coil, moves ~115Å from its initial position between subunits in the pre-fusion conformation, allowing DII domains to reposition. None of the post-fusion HRA intersubunit coiled-coil contacts are observed in F-GCNt. Instead they are replaced by two sets of 6-helix rings at the DIII interfaces. For the HRA coiled coil to form, DIII must rotate and collapse inwards, further compacting the head. The conformational change also requires the opening and translocation of the HRB stalk. In the pre-fusion form, HRB is located at the base of the head region. During the conversion to the post-fusion conformation, HRB segments must separate and swing around the base of the head, to pack against the HRA coiled coil. In the prefusion conformation, HRA is broken up into 4 helices, 2 b-strands and 5 loop, kink or turn segments. Thus, the conformational changes in HRA involve the refolding of 11 distinct segments into a single, extended a-helical conformation. The pre-fusion F structure provides a model for the stepwise induction of membrane fusion by paramyxoviruses and reveals how multiple sequence elements play distinct structural roles in the pre- and post-fusion conformations. We also solved the atomic structure of the tetrameric paramyxovirus HN protein to a resolution of 2. 8 Å, in collaboration with Theodore Jardetzky. Based on the HN structure we propose a model for HN involvement in membrane fusion that is consistent with the available data and that involves ligand-dependent changes in the HN oligomer that are driven by surface:surface interactions. In this model, the HN dimer/tetramer forms in the absence of ligands and can interact with the F protein, potentially through lateral interactions on two sides of the tetramer. Engagement of cell surface receptors could trigger the partial disassembly of the HN tetramer, assuming that the energy of binding of the individual HN sites to distinct sialic receptors is sufficient to perturb the weak NA domain interactions. Opening of the tetrameric head, driven by the energy of receptor engagement, could lead to changes in both the HN stalk region (the HN stalk domain is thought to interact with F) and the interaction with F, thus activating F for membrane fusion.
|
Enveloped Virus Assembly
We are attempting to understand the nature
of the molecular interactions that are necessary to form a virus particle.
The application of reverse genetics; i.e. the ability to rescue infectious
influenza virus and paramyxoviruses from cloned DNA, facilitates these
studies. It has been predicted that the cytoplasmic tails of the viral
integral membrane proteins will interact with internal viral components.
The nature of the protein-protein interactions is being pursued by using
reverse genetics, biochemical methods and also by using the yeast genetics
approach of interaction traps. We have shown for influenza virus that
both the hemagglutinin and neuraminidase cytoplasmic tails are a critical
determinant for forming normal virions. Similarly we have shown that the
cytoplasmic tail of HN is required for normal virus budding. We are using
an in vitro budding assay that enables us to determine the molecular requirements
for virus assembly. We are also determining the possible role of mono-ubiquitination
of viral proteins in correct assembly of cellular protein complexes that
may mediate viral budding.
Some aspects of research in our laboratory on influenza virus and paramyxoviruses are supported by grants from the National Institutes of Health.
RESEARCH
LINKS (back
to top)
Research-Related (back to top)
American Society for Virology (ASV)
American Association for the Advancement of Science (AAAS)
CMS Molecular Biology Resource
Northwestern University Index of Electronic Resources
Howard Hughes Medical Institute
Pedro's BioMolecular Research Tools
Society for General Microbiology
Journals (back to top)
Journal of Biological Chemistry
Microbiology and Molecular Biology Reviews (mmbr)
Molecular and Cellular Biology
Funding (back to top)
Research Opportunities and Funding for Northwestern University Undergraduates
Northwestern University Office of Research and Sponsored Programs
Science (AAS) Where to Search for Funding
UT Southwestern Grants Management: Funding Announcements
Specific Granting Agencies (back to top)
Damon Runyon Postdoctoral Fellowship
Howard Hughes Medical Institute
Human Frontier Science Program (fellowships to work in foreign laboratories)
Leukemia Society National Institutes of Health Grants
National Institutes of Health Grants
The National Academies Fellowships Listings
Chateaubriand Fellowship for Predoctoral Research in France
Travel Grants (Robert H. Lurie Comprehensive Cancer Center of Northwestern University) (back to top)
Katten Muchin Rosenman Travel Scholarship Program
Center for Genetic Medicine Travel Fellowship
Cancer Prevention and Control Travel Scholarship Program