Carthew Lab: Research
Gene Silencing by RNA
The genes in our genomes are usually considered “good for us” and therefore conserved and carefully expressed through our lifetimes and the lifetimes of future generations. Yet paradoxically, an ancient biochemical mechanism exists to specifically silence gene expression. This system relies upon RNA to guide the silencing. Why is there such a system and how does it work? The answer to the first question lies in our genomes, which are chock full of genes that propagate themselves at our expense. Richard Dawkins used the concept of selfish gene to describe how such genes can come into conflict with their hosts. Mechanistically, such genes propagate by being part of transposons, sequences of DNA that can move to new positions in the genome of a single cell. Transposons are found throughout the living world, and they propagate from one organism to the next by organism reproduction. At least 45% of the human genome is derived from transposons, and their jumping around the genome is a major cause of mutagenesis. Remarkably, another type of mobile genetic element is highly related to transposons: viruses. Viruses share sequence features with transposons, yet they propagate independent of the genome and they are transmitted from one organism to the next through infection.
RNA and Argonautes
Are there conserved ways that organisms fight against these mobile elements? The answer is yes. The Achilles heel of these elements lies in their autonomy of replication and transcription. This autonomy leads to transcription of both strands of coding nucleic acid into RNA, which can basepair to form an RNA double helix. Double-stranded RNA (dsRNA) associates with a protein of the Argonaute family, members of whom are found throughout the eukaryote phylogenetic tree. Even though Argonautes bind to dsRNA, they have the ability to cleave the phosphodiester backbone of one RNA strand of the paired duplex RNA. We even know which amino acids within the catalytic site of an Argonaute protein are essential for RNA cleavage (Kim et al., 2007). When the RNA strand is cleaved, it is rapidly unwound from the intact RNA strand and released from the Argonaute. The act of cleavage destroys the integrity of the RNA, rendering it unstable and unable to complete its function. This is a key mechanism to silence gene expression.
Single stranded RNA also has the ability to associate with an Argonaute protein but it will only do so with high efficiency if the Argonaute contains a complementary single-stranded RNA. This sets up a cycle of pairing-cleavage-unpairing that makes Argonaute act like a multi-turnover enzyme, specifically programmed by the single-stranded guide RNA it is stably associated with. It imbues Argonaute with awesome powers of gene silencing. The fidelity of RNA basepairing means the chance of accidentally cleaving the wrong RNA is less than 10-9. Argonaute not only cleaves RNA from the original gene from which it was derived but also RNA from all gene copies with similar sequence. Finally, the system adapts to mutation in gene sequence, because it is constantly sampling new dsRNA. So anytime there is a sequence change, the changed RNA get placed into Argonaute and silences more copies of itself.
How powerful is this RNA-Argonaute system to combat transposons and viruses? We have found that it works as a powerful antiviral immune system in Drosophila, reducing both virus load and pathogenicity (Wang et al., 2006). Many animals and most plants rely on it as a major immune response to viral pathogens. How efficacious it is depends on the host species and type of virus - and a major goal is to engineer the system to work robustly on more disease pathogens. We have also found the system works against transposons in Drosophila, reducing expression and propagation (Marques et al., 2010). Two flavors of RNA carry out transposon silencing: siRNAs and piRNAs. The former primarily silences transposons in somatic cells of the body, and they also silence viruses. The latter RNA primarily silences transposons in the sperm and egg cells of the body. Special Argonautes collaborate with siRNAs and piRNAs (Pham et al., 2004 and Kennerdell et al., 2002).
Engineered Silencing for Health and Biotechnology
If any dsRNA can associate with an Argonaute, it will silence its corresponding gene. The mechanism does not pick and choose according to whether a gene really is mobile or not. Therefore, we can program Argonautes according to our own purposes. This is done by synthesizing dsRNA and getting it into cells - once inside it associates with Argonaute and silences the targeted gene. This was first shown by Andrew Fire and Craig Mello for genes in the nematode Caenorhabditis elegans (Fire et al., 1998), and later by our group for genes in Drosophila (Kennerdell and Carthew, 1998 ). The method has grown to become an essential biomedical tool for genetic analysis on a genome-wide scale. It is also being developed by the pharmaceutical industry for treatment of diseases as wide-ranging as liver disease, cancer, and viral infection. The biotechnology sector is developing it to make agricultural crops with enhanced nutrition and pest resistance, and reduced toxicity and allergenicity.
Dicing the RNA
Transposons and viruses make dsRNA, but the RNA is originally quite diverse in its overall structure. How does Argonaute recognize this diversity? It turns out that Argonaute does not. Instead, a ribonuclease called Dicer chops the original dsRNA into small “bits” that are 20 to 23 nucleotides in length. Some species such as Drosophila have more than one type of Dicer, each with a specialized function (Lee et al., 2004). In order to chop up dsRNA, Dicer requires a partner protein with domains that bind to the dsRNA. In Drosophila, this partner protein is called Loquacious (Marques et al., 2010). Dicer not only chops dsRNA into short siRNAs but it also helps siRNAs associate with an Argonaute protein (Pham et al., 2004). In this activity, Dicer is aided by a partner protein that can sometimes be different from the dicing partner protein (Marques et al., 2010).
The Future
Our objective is a full understanding of the silencing mechanism to enhance its potential for biotechnology, disease treatment, and genome integrity. One research aim is to solve the puzzle of virus defense. Mammals have an siRNA-based silencing mechanism useful against genes but not against viruses. Why are some animal species able to use the siRNA-based mechanism for virus defense but other species, including humans, lack this ability? Can we devise ways to engage the mammalian mechanism against viruses? Another research aim is to understand how piRNAs get made. They are not made by any Dicer enzyme but use a different mechanism to process transposon dsRNA into piRNAs. A third research aim is to determine why there is an intimate relationship between siRNA-based silencing and the sorting of membrane cargo within the cytoplasm. We discovered such a relationship with cargo that is trafficked through endosomes to lysosome-related organelles (Lee et al., 2009 and Harris et al., 2011 ). Is the relationship due to the way in which viruses often enter cells through the endocytic pathway or for some other reason? The reason why this relationship exists is tantalizingly mysterious but the fact that it is conserved between Drosophila and humans argues that the reason is of general importance.
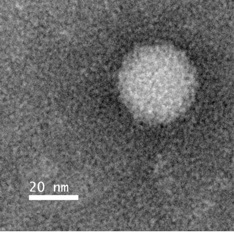
Image of a picorna-like virus from Drosophila that is inhibited by the siRNA-based mechanism operating in flies. Picornaviruses in humans include polio and hepatitis A virus.
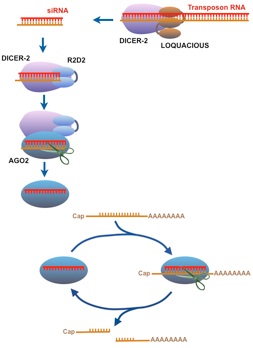
The siRNA-based mechanism in Drosophila. Double helical RNA is chopped by Dicer-2 with help from Loquacious to form siRNAs. Released siRNA associates with a protein dimer complex of Dicer-2 and R2D2. The RNA-protein complex recruits an Argonaute called Ago2, and the siRNA is transferred to Ago2. One RNA strand is cleaved and released, leaving the other siRNA strand to act as a guide for Ago2. If a mRNA transcript can basepair with the guide RNA, then Ago2 cleaves the transcript and releases it. The cleaved RNA is disabled and Ago2 is free to seek and destroy more copies of the transcript.

Northwestern University | Northwestern Search
Center for Cell and Developmental Systems Biology | Department of Molecular Biosciences
2205 Tech Drive, Hogan 2-100, Evanston, IL 60208 Email the Webmaster
WWW Disclaimer and University Policy Statements © 2011 Northwestern University